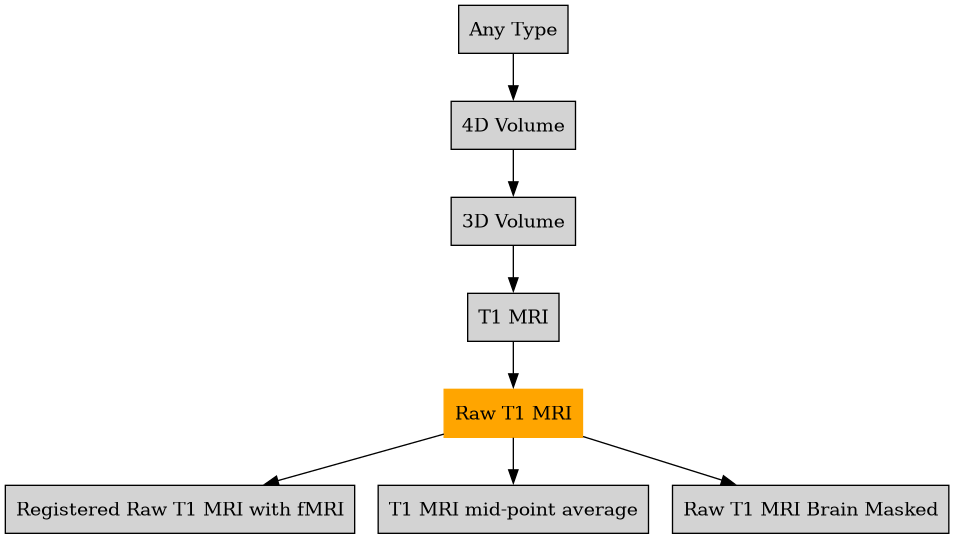

Defined in file : brainvisa/types/builtin.py
01 Create Freesurfer subject from T1 anatomical image
18 Meshes normalization
AC/PC Or Normalization
Add Border To Image
Add gaussian noise in image background
Ana Brain Mask from T1 MRI
Anatomist Show Bias Correction
Anatomist show Commissure coordinates
Anatomist Show White Ridge on T1 MRI
Anatomy Normalization (using AimsMIRegister)
Anatomy Normalization (using Baladin)
Anatomy Normalization (using FSL)
Anatomy Normalization (using FSL) with Re-initialization
Anatomy Normalization (using SPM)
Anatomy Normalization (using SPM) with Re-initialization
Anatomy Normalization (using SPM12)
Anatomy Normalization (using SPM12) with Re-initialization
Anatomy Normalization (using SPM8)
Anatomy Normalization (using SPM8) with Re-initialization
Baladin Normalization Pipeline
CAT12 - Segment
Check raw data pipeline (PET)
Complete T1 MRI acquisition data
convert Dicom to Nii
Correction Brain Mask from T1 MRI
disco_subject_to_icbm
Freesurfer / BrainVisa full pipeline
FSL Normalization Pipeline
Import Baby T2 MRI
Import Brain Mask
Import Dicom T1 MRI
Import Ecat PET Dynamic & T1MRI
Import FreeSurfer grey/white segmentation to Morphologist
Import MCRIBS dHCP preterm subject into a brainvisa database
Import MNI CIVET Segmentation
Import T1 MRI
Import T1 MRI, longitudinal version
Import T1MRI (nifti) about PET subject
Import volBrain processes results
Import volBrain results
Label volume editor
Localize Talairach Coordinate
morpho_report
Morphologist 2011
Morphologist 2012
Morphologist 2013
Morphologist 2015
Morphologist 2025
Normalization pipeline
Optimal PET Amyloid Long (OPAL spm12)
pet pipeline using MRI (spm12)
pet pipeline using MRI (spm8)
Pipeline Baby
Pipeline Baby (Utrecht)
Prepare Subject for Anatomical Pipeline
Preprocess for PET Pipeline using MRI (spm12)
Preprocess for PET Pipeline using MRI (spm8)
Read and complete AC-PC files
Reorient Anatomy
Run AssemblyNet
segmentation choice (spm8)
Segmentation of Cerebellum, Central Grey Nuclei and Ventricules (Utrecht)
set_image_normalization
Simplified Morphologist 2025
Skull-stripped brain normalization
Skull-stripping
SPM Normalization Pipeline
spm12 - Segment
spm8 - New Segment
spm8 - VBM Segmentation
Sulcal Lines Extraction Left
Sulcal Lines Extraction Right
T1 Bias Correction
T1 Bias Correction
T1 mapping maps: build using VFA method
T1 Pipeline 2007
Talairach Transformation From Normalization
Transform a commissures AC-PC coordinates
Validation Pipeline
Validation_1 Bias Correction from T1 MRI
View in hippocampic referential
Aperio svs
BrainVISA volume formats
DICOM image
Directory
ECAT i image
ECAT v image
FDF image
FreesurferMGH
FreesurferMGZ
GIS image
gz compressed ECAT i image
gz compressed ECAT v image
gz compressed GIS image
gz compressed MINC image
gz compressed NIFTI-1 image
gz compressed SPM image
Hamamatsu ndpi
Hamamatsu vms
Hamamatsu vmu
JPEG image
Leica scn
MINC image
NIFTI-1 image
Sakura svslide
SPM image
TIFF image
TIFF(.tif) image
Ventana bif
Z compressed ECAT i image
Z compressed ECAT v image
Z compressed GIS image
Z compressed SPM image
Zeiss czi
{protocol}/{subject}/anatomy/<subject>
{protocol}/{subject}/anatomy/w<subject>
{protocol}/{subject}/anatomy/<subject>_*
{protocol}/{subject}/anatomy/n<subject>_*
{protocol}/{subject}/anatomy/{acquisition}/<subject>
{protocol}/{subject}/anatomy/{acquisition}/w<subject>
{protocol}/{subject}/anatomy/{acquisition}/<subject>_*
{protocol}/{subject}/anatomy/{acquisition}/n<subject>_*
{protocol}/{subject}/t1mri/{acquisition}/<subject>
{protocol}/{subject}/t1mri/{acquisition}/normalized_{normalization}_<subject>
{center}/{subject}/t1mri/{acquisition}/<subject>
{center}/{subject}/t1mri/{acquisition}/normalized_{normalization}_<subject>
<filename>_t1
normalized_<filename>
sub-{subject}/ses-{session}/anat/t1mri/{acquisition}/<subject>
sub-{subject}/ses-{session}/anat/t1mri/{acquisition}/normalized_{normalization}_<subject>
sub-{subject}/ses-{session}/{bids}/sub-<subject>_ses-<session>_<bids>_desc-conform_T1w
sub-{subject}/ses-{session}/{bids}/sub-<subject>_ses-<session>_<bids>_normalized_{normalization}
sub-{subject}/ses-{session}/anat/sub-<subject>_ses-<session>_{bids}_T1w
sub-{subject}/ses-{session}/anat/sub-<subject>_ses-<session>_T1w
sub-{subject}/ses-{session}/anat/sub-<subject>_{bids}_T1w
sub-{subject}/ses-{session}/anat/sub-<subject>_ses-<session>_{bids}
sub-{subject}/ses-{session}/anat/sub-<subject>_ses-<session>
sub-{subject}/ses-{session}/anat/sub-<subject>_{bids}
sub-{subject}/anat/sub-<subject>_ses-{session}_{bids}_T1w
sub-{subject}/anat/sub-<subject>_ses-{session}_T1w
sub-{subject}/anat/sub-<subject>_{bids}_T1w
sub-{subject}/anat/sub-<subject>_ses-{session}_{bids}
sub-{subject}/anat/sub-<subject>_ses-{session}
sub-{subject}/anat/sub-<subject>_{bids}
protocol , subject , spm_normalized , filename_variable , acquisition
protocol , subject , acquisition , normalization
rescan , time_duration , time_point , acquisition_date , center , subject , acquisition , normalization , acquisition_sequence , analysis , processing
rescan , time_duration , time_point , acquisition_date , subject , session , acquisition , normalization , analysis
rescan , time_duration , time_point , acquisition_date , subject , session , bids , normalization , analysis
subject , session , bids